Chapter | 2 Analyses plots
2.1 UpSet plot of merged analyses sources
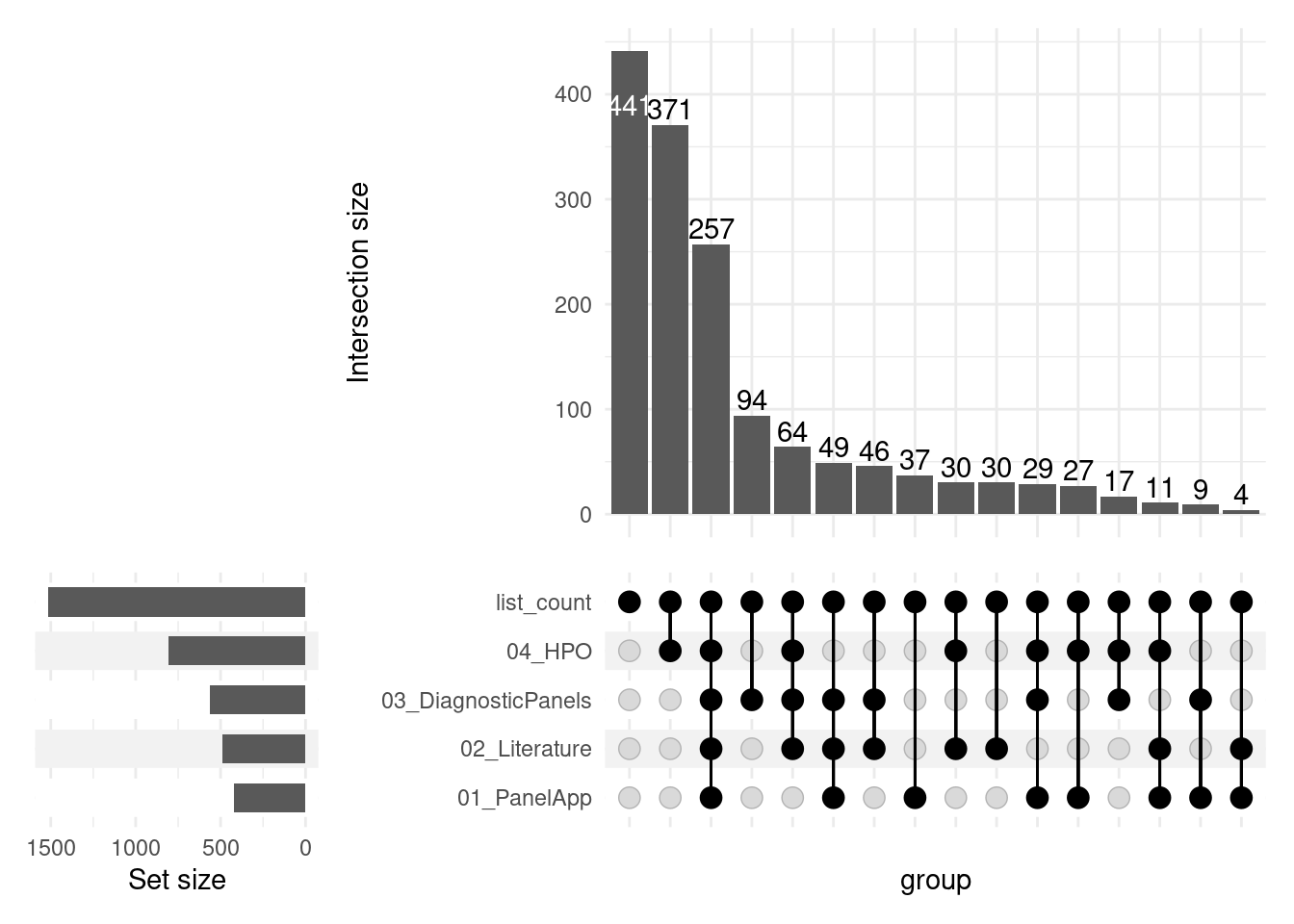
Below you can see a UpSet plot of the merged analyses.
In the lower left corner you can see the number of Genes originating from each of the different resources, after that resources are sorted on the right side. UpSet plots generally represent the intersections of a data set in the form of a matrix, as can be seen in the graph below.
- Each column corresponds to a set, and the bar graphs at the top show the size of the set.
- Each row corresponds to a possible intersection: the dark filled circles show which set is part of an intersection.
- For example, the first column shows that most of the genes found in only one of the five sources are derived from the PubTator query, and in the third column you can see that 177 Genes are found in all five sources.
2.2 Bar plot of PanelApp results
Below you can see a Bar plot of the PanelApp analysis.
We retrieved all kidney disease related panels from both PanelApp UK and PanelApp Australia, meaning all panels that include “renal” or “kidney” in its name.
- The y axis shows the number of Genes in the different panels, which is also visualized by the height of the bars.
- The x axis displays the number of panels (source_count), i.e. in how many different panels a single Gene occurred.
- For example 38 Genes occurred in just one panel and 2 Genes were present in all thirty different panels.
2.3 Bar plot of Literature results
Below you can see a Bar plot of the Literature analysis.
We identified Genes associated with kidney disease in a systematic Literature search using the following search query:
(1) “Kidney”[Mesh] OR “Kidney Diseases”[Mesh] OR kidney OR renal AND
(2) “Genetic Structures”[Mesh] OR “Genes”[Mesh] OR genetic test OR gene panel OR gene panels OR multigene panel OR targeted panel*
- The y axis shows the number of Genes in different publications, which is also visualized by the height of the bars.
- The x axis displays the number of publications (source_count), i.e. in how many different publications a single Gene occurred.
- For example 331 Genes occurred in just one of the publications and 1 Gene was present in all 13 different publications.
2.4 Bar plot of Diagnostic panels results
Below you can see a Bar plot of the Diagnostic panels analysis.
We used ten common diagnostic panels that can be purchased for genome analysis and extracted the screened Genes from them.
- The y axis shows the number of Genes in the different diagnostic panels, which is also visualized by the height of the bars.
- The x axis displays the number of panels (source_count), i.e. in how many different panels a single Gene occurred.
- For example 371 Genes occurred in just one panel and 56 Genes were present in all ten different panels.
2.5 Bar plot of HPO in rare disease databases results
Below you can see a Bar plot of the HPO-term based query in rare disease databases (OMIM, Orphanet).
We used eight common databases for rare diseases and screened them for kidney disease associated Genes from a Human Phenotype Ontology (HPO) based search query. The most comprehensive HPO term used was “Abnormality of the upper urinary tract” (HP:0010935) and included all sub group terms. We deliberately chose these to be somewhat broader in order to fully include all relevant kidney diseases such as CAKUT, among others.
- The y axis shows the number of Genes in the different rare disease databases, which is also visualized by the height of the bars.
- The x axis displays the number of databases (source_count), i.e. in how many different databases a single Gene occurred.
- For example 652 Genes occurred in just one database and 1 Gene was present in all eight different databases.
2.6 Bar plot of PubTator results
Below you can see a Bar plot of the PubTator analysis.
We retrieved all kidney disease associated Genes from a PubTator API-based automated literature extraction of publications available on PubMed.
- The y axis shows the number of Genes in the different publications, which is also visualized by the height of the bars.
- The x axis displays the number of publications (source_count), i.e. in how many different publications a single Gene occurred.
- For example 914 Genes occurred in just one publication and 1 Gene was present in 1221 different publications.